Biotech platform under the hood
Bioinformatics is a young science, for most of the IT community a foreign and magical discipline that seems like something out of a science fiction book. Therefore, the banal explanation that "a bioinformatics platform is a product that enables scientists in this field to collaborate more easily and make new discoveries faster" does not reveal much.
In this text, we will try to shed light on at least part of that mystery for you, because very soon, when we scratch below the surface, we find ourselves on much more familiar terrain of the classic problems that computer science deals with.
A brief history of bioinformatics
But before that, a very brief overview of important things from history and the reason why this discipline has flourished recently and attracted large companies that invest in its development.
Genetics, the science that studies genes, is not that new. Gregor Mendel experimented with peas almost 2 centuries ago, but a holistic view of the entire human genome was unreachable to us until the early 2000s. It was then that almost the entire genome was sequenced for the first time in the Human Genome Project. The long sequence of about 3 billion base pairs (ACTG) that make up the human DNA chain has been mapped to groups of over 20,000 genes.
That project was fabulously expensive, but the progress in various sciences (from molecular biology to computer science) has led to the fact that in the last ten years, it is possible to read the genome of one person for an affordable sum of money, from a few hundred to a few thousand dollars. Given that such analyzes are already being used in medicine, and have enormous potential for the future, and are the subject of numerous cutting-edge researches, it is natural that such readings (sequencing) have become more frequent and data has begun to be generated with exponential growth.
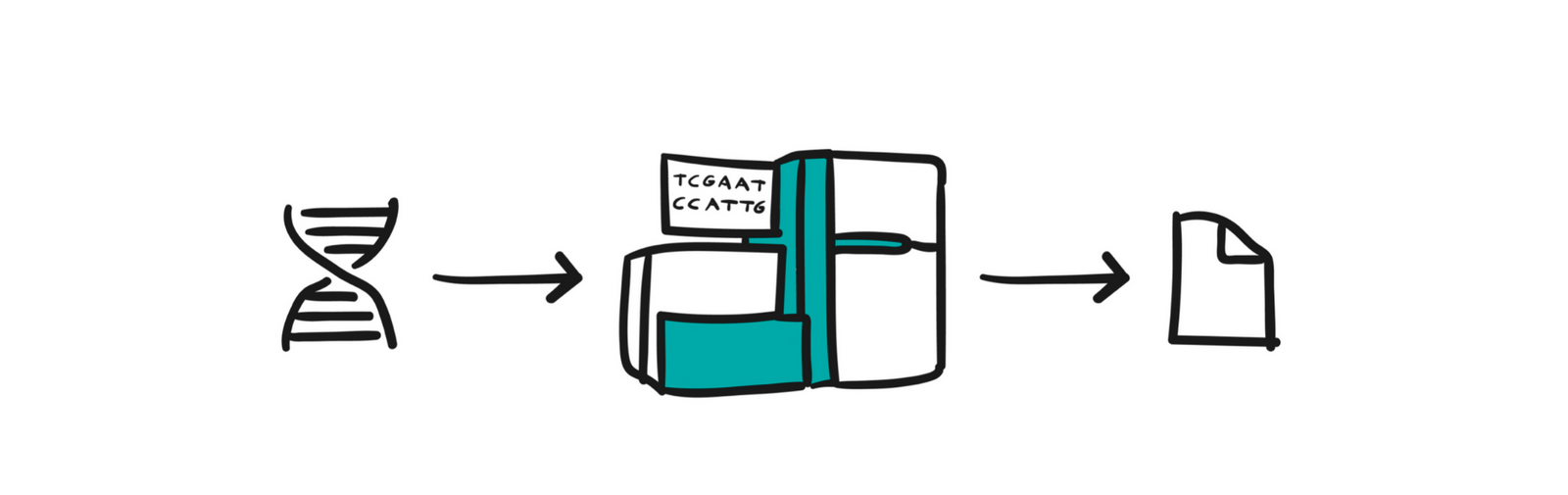
Now it is important to note that the result of sequencing one sample is a text file of a size of several hundred GB, and it is precisely this jump in the amount of information that we want to process that gave rise to the need for the development of bioinformatics. In this field, although it can still be said that it is at an early stage, we are dealing with petabytes of genomic data, and in the not-too-distant future exabytes await us.
So what is a bioinformatics platform?
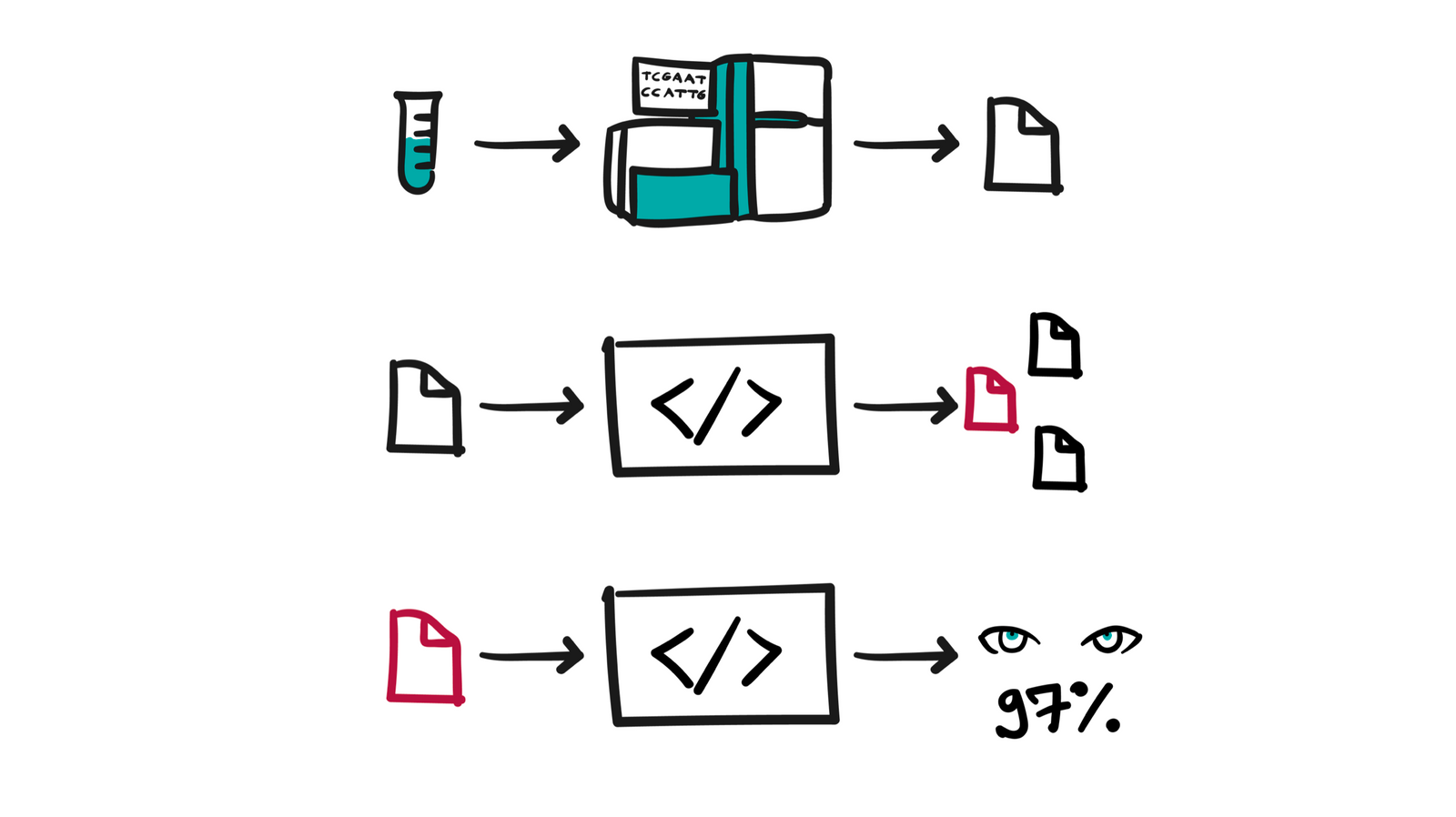
The job of the bioinformatics platform is to facilitate the analysis of this large amount of data, to do it in a safe manner following strict safety standards - because we are talking about data of an extremely sensitive nature, to enable the repeatability and verifiability of analyzes that are essential for any scientific research, search and visualization of results processing and to provide the scalability needed for the smooth operation of multiple clients on one environment.
The core of a bioinformatics platform is certainly computation - support for the execution of diverse analyses, which are actually a branched sequence of different tools from the domain of bioinformatics, performed on a set of data and configurations. Bioinformatics analyses are demanding, because they work on large amounts of data, but they are often very complex because they can consist of a large number of interdependent steps that are described by some directed execution graph. Bioinformatics tools require a lot of processing power and other resources (memory/disk/network), so the platform is responsible for orchestrating the execution of this work. The platform on behalf of the user dynamically requests tens of thousands of CPU cores from the Cloud Provider needed to perform some set of analyses, and uses them for this purpose for a few hours or a few days, as long as it takes to complete them, and finally deallocates all the rented resources, saving only the final processing results.
If we look from a little more distance, we can look at this aspect of the platform as a standard MapReduce system, modified and specialized for the needs of the domain in which it works, but still completely limited by the laws of distributed systems much more familiar to the IT community. Work on optimizing the efficiency and scalability of this part of the system is an endless source of purely computational challenges.
The development of Cloud technologies was an essential trigger that enabled the practicality and efficiency of the platform, it allows us to manage resources of exactly the right size only as much as we need and thus provides resistance to the variety of analyzes that are performed. Cloud Computing today is easy to use and very comfortable on a small scale, but on a large scale, it becomes complicated and brings with it specific challenges of use. Cloud providers are not used to such abrupt changes in demands and use of various resources, where within a few tens of minutes we increase the use a few thousand times compared to what we need during "quiet" periods. Keeping this part of the system stability is a discipline unto itself. If we include in the story the fact that our clients themselves are users of different cloud technologies from several vendors (AWS, Google, Azure, Aliyun...) to which they are connected, the platform's job is to reconcile and unify the possibilities and offer comfort and uniform quality of work, whichever vendor the user chooses.
Nothing without big data
The results of individual analyzes are relevant to the individual case and unlock different applications with a view to the individual. On the other hand, there is a great need to process the data of a larger group of people (cohorts) who exhibit characteristics that are interesting for some research. When we put into perspective the raw amount of bytes associated with one individual, cohort analysis requires the application of different research techniques for the simple reason of the limitations of today's technologies, in cases where we are dealing with tens or hundreds of terabytes of "live" data from which we want to draw some conclusion, ideally in the shortest period of time and as cheaply as possible.
For such research, it is necessary to provide users with several different interfaces, programmatic and visual, through which it is possible to experiment live with the data and quickly check different hypotheses. Here we are stepping into the ever-popular BigData terrain, where knowledge of optimization techniques and new technologies is imperative to successfully tackle this cross-domain problem.

Is that all?
In the end, the platform is not only a tool for processing data at the micro and macro level, but also an ecosystem. It is necessary to provide all those things that make a product complete. Possibilities for the collaboration of several scientists/researchers, sharing and publication of works and results, developing and testing new tools, monitoring analysis metrics, charging resources, maintaining access rights, and enabling comfortable and comprehensive programmatic access to the platform, there are dozens of products integrated into one whole.
On the other side of the firewall, the one that is not visible to the users, there is the monitoring of the complex architecture made up of hundreds of microservices, the struggle with the security requirements that permeate the entire system, the improvement of performance to support new clients and completely different and innovative ways of processing data.
The bottom line is that although the domain is what makes our work interesting, it is largely due to the very nature of the work and the problems that the domain brings. You don't have to understand genomics, it is not necessary (although it is useful) to be interested in biology - if you like distributed systems, complex architectures, algorithmic problems, data science, cloud computing, query optimization over databases, complex frontend visualizations, improving engineering skills through methodical work, you will surely find some part of the platform where you will be able to express your talent, do something that fulfills you and help us code together for a healthier tomorrow.
See all open positions at Seven Bridges at this link.


